|
DriverMap AIR TCR-BCR (CDR3 only) Profiling
|
DriverMap AIR TCR-BCR (Full-Length Variable Region) Profiling
|
|
Region
|
CDR3 only
|
FR1, FR2, FR3, CDR1, CDR2 and CDR3
|
|
Amplicon Length (bp)
|
~ 300 bp
|
~ 600 bp
|
|
Primer Location
|

|

|
|
Starting Material
|
RNA or DNA
|
RNA
|
|
Model System
|
Human (RNA or DNA)/ Mouse (RNA)
|
Human
|
|
NGS Instrument Options
|
Illumina NextSeq500/550, NextSeq 2000, NovaSeq
|
Illumina NextSeq 2000 (x 2 300 cycles) kit only
|
|
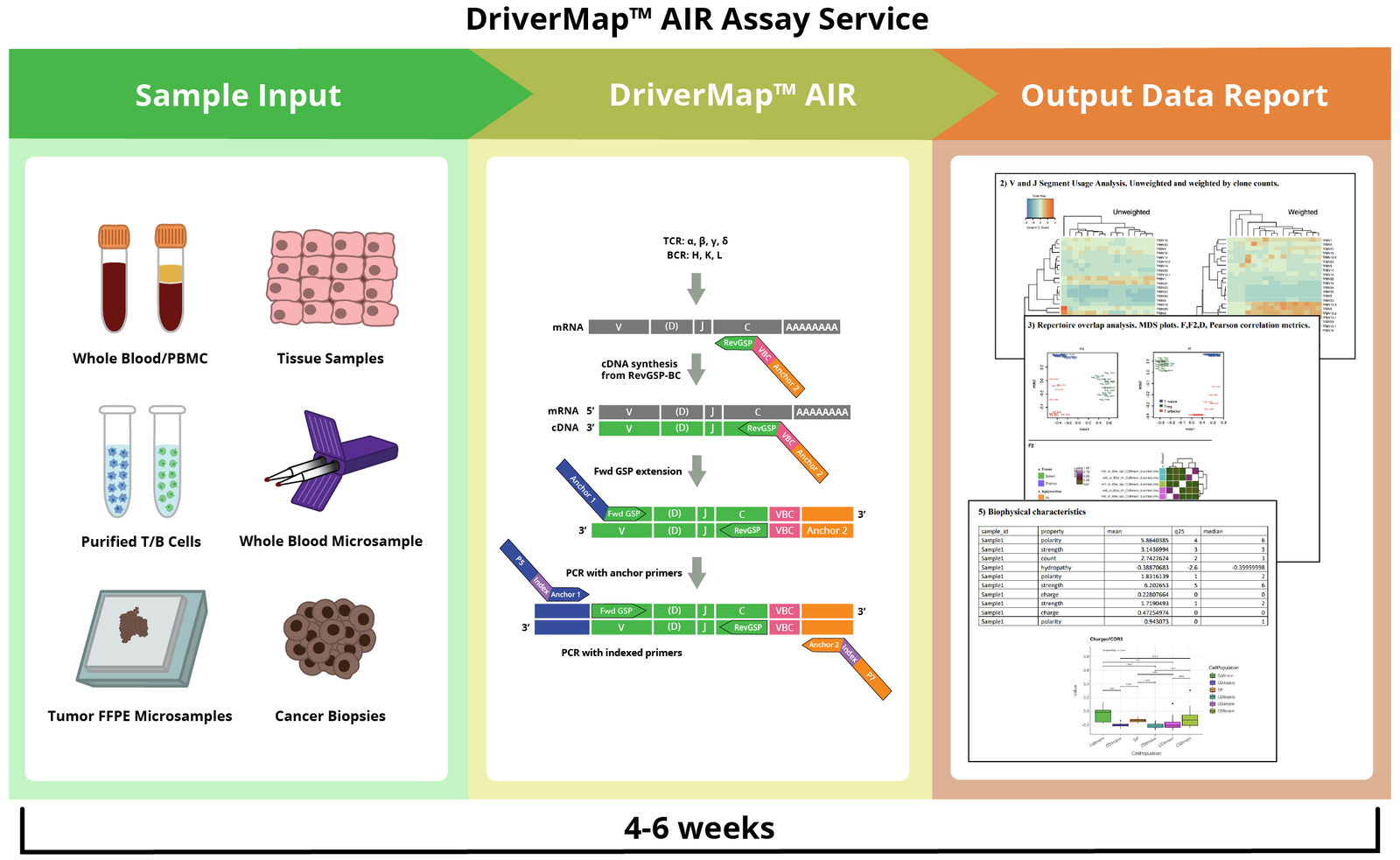
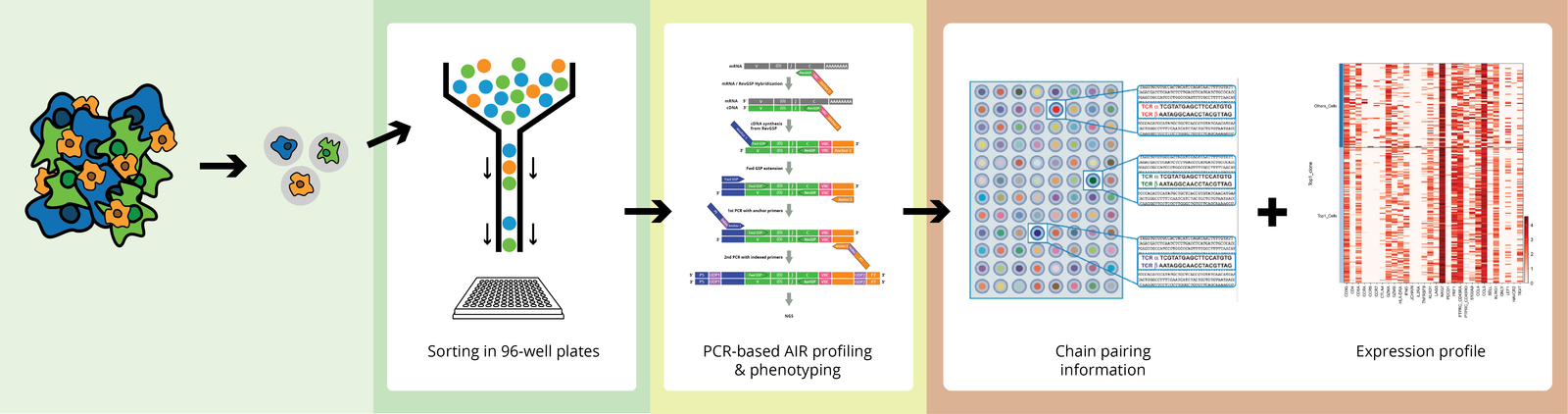
How it Works
|
The CDR3 region determines the structure and specificity of the antigen. The majority of repertoire profiling studies are based on the analysis of only the CDR3 region, as this is the hypervariable region and contains crucial information to study immune repertoire diversity.
|
The V region encodes CDR1 and CDR2 in germline DNA segments and primarily interacts with the major histocompatibility complex (MHC) molecules. The full-length receptor region is involved in antigen receptor binding affinity and/or downstream signaling and allows one to clone and express the identified and chosen receptors.
|
|
Key Applications
|
-
Tracking T cell diversity and clonality
-
Finding markers of autoimmune diseases
-
Studying immune cell migration patterns
|
|


























 U.S. List Price
U.S. List Price